فروشگاه
Showing 321–340 of 605 results
فیلتر ها-
پروبهای فیش
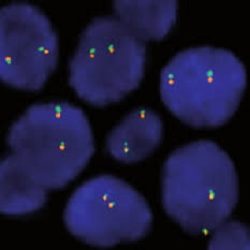
پروب فیش SS18 Break Apart FISH Probe
Rated 0 out of 5Read moreINTRODUCTION: Abnormal expression of the SS18 gene (also known as SYT or SSXT) has been described in up to 80% of synovial sarcomas (regardless of their morphology) in which SYT-SSX1, SYT-SSX2, SYT-SSX4 translocations can appear. All of them are involved in their pathogenesis, but also in the prognosis of the neoplasm. The SYT-SSX1 one shows, in general, a less favorable prognosis. Rarely, translocation t(X-18) has been detected in pleomorphic sarcomas (previously named as malignant fibrous histiocytoma) or in fibrosarcomas.
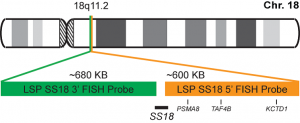
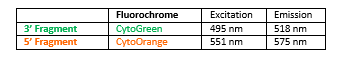
INTENDED USE: The SS18 Break Apart FISH probe is designed to detect rearrangements in the SS18 human gene located in the band of the chromosome 18q11.2. Aside from detecting breaks that can lead to translocation of parts of the gene, to its inversion or to its fusion with other genes, the probe can also be used to identify other aberrations of SS18 such as deletions or amplifications.
PROBE DESCRIPTION: The 5’ telomeric fragment of the SS18 Break Apart FISH probe labeled with the fluorochrome CytoOrange covers the 5’ portion (beginning) of the SS18 gene and some adjacent genomic sequences.The 3’ centromeric fragment of the SS18 Break apart FISH probe labeled with CytoGreen covers the 3’ part (end) and the adjacent and inferior sequences of the gene. Both probes are flanking sequences in the SS18 gene, in which variable cut-off points have been observed.

-
پروبهای فیش
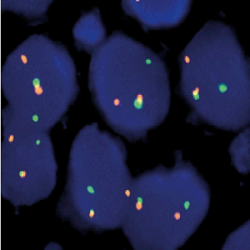
پروب فیش FOXO1 Break Apart FISH Probe
Rated 0 out of 5Read moreINTRODUCTION: Rearrangements and abnormal expression of the FOXO1 gene (also known as FKH1, FKHR or FOXO1A) have been observed in the alveolar rhabdomyosarcoma, prostate carcinoma and other types of tumors. In the case of alveolar rhabdomyosarcoma, fragments of the 3’ region of the FOXO1 gene translocate with fragments of the 5’ portions of the PAX3 and PAX7 genes, located in the chromosome 2 and 1 respectively. Those cases showing the PAX3-FOXO1 fusion in metastases present a worse prognosis, while in primary tumors, the development of the disease is independent of the type of translocation. In the prostate cancer, deletions of the FOXO1 gene have been detected in up to 30% of the cases.
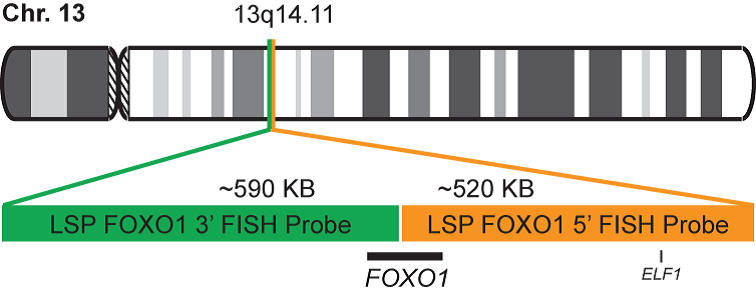
INTENDED USE: The FOXO1 Break Apart FISH probe is designed to detect rearrangements in the FOXO1 human gene located in the band of the chromosome 13q14.11. Aside from detecting breaks that can lead to translocation of parts of the gene, to its inversion or to its fusion with other genes, the probe can also be used to identify other aberrations of the FOXO1 gene such as deletions or amplifications.
PROBE DESCRIPTION: The 5’ telomeric fragment of the FOXO1 Break Apart FISH probe labeled with the fluorochrome CytoOrange covers the center and the 5’ portion (beginning) of the FOXO1 gene and some adjacent genomic sequences. The 3’ centromeric fragment of the FOXO1 Break apart FISH probe labeled with CytoGreen covers the 3’ part (end) of the FOXO1 gene as well as the adjacent and inferior sequences of the gene. Both probes are flanking sequences of the FOXO1 gene, in which variable cut-off points have been observed.
-
پروبهای فیش
پروب فیش TFE3 Break Apart FISH Probe
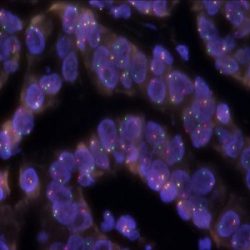
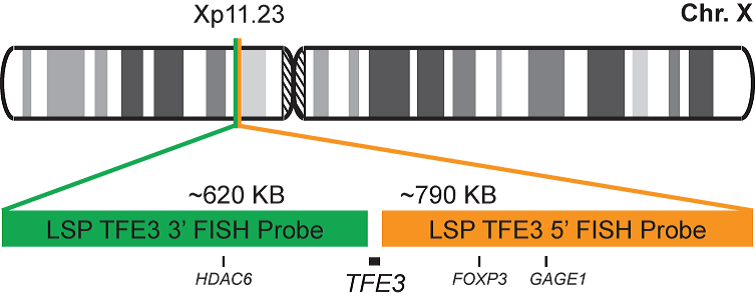
Rated 0 out of 5Read moreINTRODUCTION: Rearrangements and abnormal expression of the TFE3 gene (also known as TFEA, RCCP2, RCCX1 or bHLHe33) have been described in a special subtype of renal cell carcinoma associated with Xp11.2 translocation, in alveolar sarcoma and other tumor types.
INTENDED USE: The TFE3 Break Apart FISH probe is designed to detect rearrangements in the TFE3 human gene located in the band of the chromosome Xp11.23. Aside from detecting breaks that can lead to translocation of parts of the gene, to its inversion or to its fusion with other genes, the probe can also be used to identify other aberrations of TFE3 such as deletions or amplifications.
PROBE DESCRIPTION: The 5’ centromeric fragment of the TFE3 Break Apart FISH probe labeled with the fluorochrome CytoOrange covers the 5’ portion (beginning) of the TFE3 gene and some adjacent genomic sequences. The 3’ telomeric fragment of the TFE3 Break apart FISH probe labeled with CytoGreen covers the 3’ part (end) and the adjacent and inferior sequences of the gene. Both probes are flanking sequences in the TFE3 gene, in which variable cut-off points have been observed.
-
پروبهای فیش
پروب فیش DDIT3 (CHOP) Break Apart FISH Probe
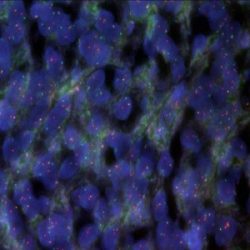
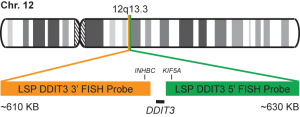
Rated 0 out of 5Read moreINTRODUCTION: Mixoid / round cell liposarcoma (MRCLS) is the most common liposarcoma. In comparison with other liposarcomas, MRCLS has a greater tendency to metastasize. Rearrangements and abnormal expression of the DDIT3 gene also known as CHOP, CEBPZ, CHOP10, CHOP-10, GADD153 in myxoid liposarcoma and other malignancies have been observed. One most common abnormality is the reciprocal translocation t (12; 16) (q13; p11) that is present in up to 95% of cases. The resulting FUS-DDIT3 fusion protein has oncogenic potential and interferes with adipogenic differentiation. EWSR1 is an alternative translocation partner of DDIT3 and the resulting fusion protein EWSR1-DDIT3, which originates at t (12; 22) (q13; q12), accounts for <5% of MRCLS cases. The DDIT3 rearrangements are highly specific for MRCLS due to the presence of the FUS-DDIT3 or EWSR1-DDIT3 fusion genes.
INTENDED USE: The DDIT3 Break Apart FISH probe is designed to detect rearrangements in the human DDIT3 gene located in the band of chromosome 12q13.3. In addition to detecting breaks that can lead to the translocation of parts of the gene, its inversion or its fusion to other genes, the probe can also be used to identify other aberrations of the DDIT3 gene such as deletions or amplifications.
PROBE DESCRIPTION: The telomeric 5 ‘fragment of the DDIT3 Break Apart FISH probe labeled with the CytoGreen fluorochrome covers some genomic sequences adjacent to the 5′ end of the DDIT3 geneThe 3’-centromeric fragment of the DDIT3 Break Apart FISH probe labeled with CytoOrange covers some genomic sequences of the 3 ‘end of the gene.The two probes are sequences that flank the DDIT3 gene region where several cut points have been observed.

-
پروبهای فیش
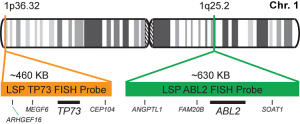
پروب فیش 1p36/1q25 FISH Probe
Rated 0 out of 5Read moreINTRODUCTION: Deletions and other abnormalities of the regions 1p and 19q have been observed in gliomas, astrocytomas, and other malignant diseases. The detection of the 1p/19q co-deletions represents a diagnostic criterion for oligodendrogliomas.
INTENDED USE: The FISH 1p36/1q25 probe is designed to simultaneously detect changes in the human TP73 and ABL2 genes, located in the 1p36.32 and 1q25.2 chromosome bands, respectively.
PROBE DESCRIPTION: The 1p36 FISH probe labeled with the CytoOrange fluorochrome covers a chromosome region that includes the whole TP73 gene located in the telomere region of the short arm of chromosome 1 and represents the target signal. The 1q25 FISH probe labeled with the CytoGreen fluorochrome covers a chromosome region that includes the whole ABL2 gene.

-
پروبهای فیش
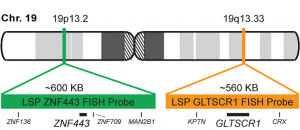
پروب فیش 19q13/19p13 FISH Probe
Rated 0 out of 5Read moreINTRODUCTION: Deletions and other abnormalities of the regions 1p and 19q have been described in gliomas, astrocytomas, and other malignant diseases. The 1p/19q co-deletions represent a diagnostic criterion in the case of oligodendrogliomas.
INTENDED USE: The FISH 19q13/19p13 is designed to simultaneously detect changes in the human GLTSCR1 and ZNF443 genes, located in the 19q13.33 and 19p13.2 chromosome bands, respectively.
PROBE DESCRIPTION: The 19q13 FISH probe labeled with the CytoOrange fluorochrome covers a chromosome region that includes the whole GLTSCR1 gene in the telomere region of the long arm of chromosome 19 and represents the target signal.The 19p13 FISH probe labeled with the CytoGreen fluorochrome covers a chromosome region that includes the whole ZNF443 gene.

-
پروبهای فیش
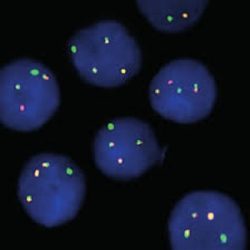
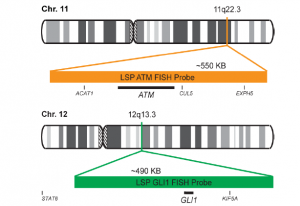
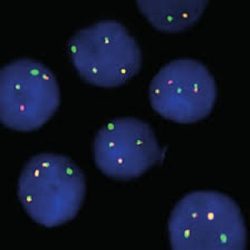
پروب فیش ATM/GLI1 FISH Probe
Rated 0 out of 5Read moreINTRODUCTION: Abnormalities in ATM – also known as AT1, ATA, ATC, ATD, ATE, ATDC, TEL1 or TELO1 – occur in ataxia telangiectasia, and have also been observed in chronic lymphocytic leukemia (CLL) and other malignancies. The GLI1 gene – also called GLI – is an oncogene that is expressed and sometimes amplified in several human cancers.
INTENDED USE: The ATM/GLI1 FISH Probe Kit is designed to detect rearrangements involving the human ATM and GLI1 genes located on chromosome bands 11q22.3 and 12q13.3, respectively.
PROBE DESCRIPTION:The ATM Probe of the ATM/GLI FISH Probe kit labeled with the fluorochrome CytoOrange covers a chromosomal region which includes the entire ATM gene. The GLI Probe of the ATM/GLI FISH Probe Kit labeled with CytoGreen covers a chromosomal region which includes the entire GLI1 gene. Both probes are designed to measure the copy number of the human ATM and GLI1 gene located on chromosome band 11q22.3 and 12q13.3, respectively.

-
پروبهای فیش
پروب فیش BCR-ABL1 Fusion/Translocation FISH Probe
Rated 0 out of 5Read moreINTRODUCTION: The product of the human BCR gene, breakpoint cluster region (BCR) protein, also called BCR1, PHL or renal carcinoma antigen NY-REN-26, is a ubiquitously expressed serine/threonine kinase and GTPase-activating cytoplasmic enzyme. ABL1 (Abelson murine leukemia viral oncogene homolog 1, also called c-ABL, JTK7, p150) is a cytoplasmic and nuclear tyrosine kinase involved in a variety of cellular processes including migration, adhesion, differentiation and apoptosis. It is also an important component of the T-cell receptor signaling system and required for T-cell development and function.
The t(9;22)(q34;q12) reciprocal translocation between BCR and ABL1 leads to a lengthened chromosome 9 (9q+) and a shortened chromosome 22 (der(22), 22-), also called Philadelphia-chromosome (Ph, Ph1). Discovered in 1960, it is a hallmark of chronic myelogenous leukemia (CML) and was the first consistent genetic abnormality identified in any human cancer.The fusion combines varying portions of the 5’- part of BCR with the 3’-part (exons 2-11) of ABL1 and leads to the production of a fusion protein in which the tyrosine kinase of ABL is constitutively activated.The Philadelphia chromosome is present in virtually all CML cases.16 However, it is also found in a subset of adult and pediatric acute lymphoblastic leukemia (ALL) and sometimes in acute myelogenous leukemia (AML) and other malignancies such as the very rare chronic neutrophilic leukemia (CNL).
INTENDED USE: The BCR-ABL1 Fusion/Translocation FISH probe kit pair is designed to detect rearrangements involving regions of the human BCR (Breakpoint cluster region) locus, located on chromosome band 22q11.23, and of the human ABL1 gene on 9q34.12. The Probe kit is optimized to reveal the typical t(9;22)(q34;q11) translocation as well as other translocations between the two genes, in metaphase and interphase blood or bone marrow cells.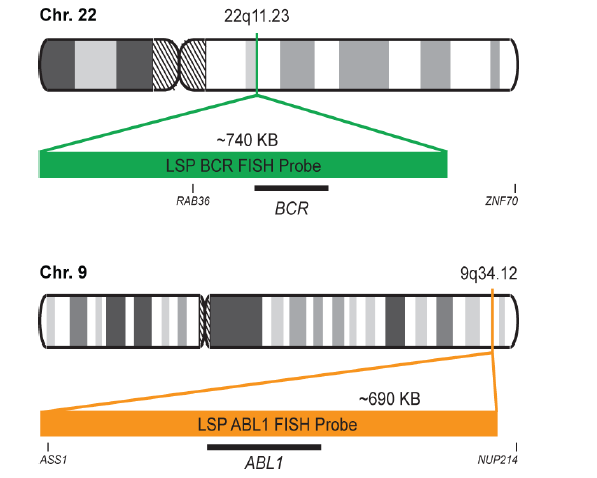
PROBE DESCRIPTION:
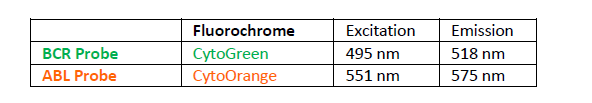
The BCR Probe of the BCR/ABL1 Fusion/Translocation FISH Probe Kit labeled with the fluorochrome CytoGreen covers the entire BCR gene along with some upstream (5’) and downstream (3’) flanking untranslated genomic sequences. The probe overlaps the known major and minor breakpoints observed in the region. The ABL1 Probe of the BCR/ABL1 Fusion/Translocation FISH Probe Kit labeled with the fluorochrome CytoOrange covers the complete ABL1 gene on 9q34.12, including the common breakpoint region upstream of exon 2, as well as adjacent 5’ and 3’ untranslated genomic sequences.
-
پروبهای فیش
پروب فیش CBFB Break Apart FISH Probe
Rated 0 out of 5Read moreINTRODUCTION:The human CBFB genes encode a protein named Core-binding factor subunit beta.
The protein encoded by this gene is the beta subunit of a heterodimeric core-binding transcription factor belonging to the PEBP2/CBF transcription factor family which master-regulates a host of genes specific to hematopoiesis (e.g., RUNX1) and osteogenesis (e.g., RUNX2).Rearrangements and abnormal expression of the CBFB gene – also known as CBFb or PEBP2B – have been observed in acute myeloid leukemia (AML) and other hematological malignancies.
INTENDED USE: The CBFB Break Apart FISH probe Kit t is designed to detect rearrangements in the human CBFB gene located on chromosome band 16q22.1. In addition to revealing breaks, which can lead to translocation of parts of the gene, inversion, or its fusion to other genes, the probe set can also be used to identify other CBFB aberrations such as deletions or amplifications.PROBE DESCRIPTION:The 5’ centromeric fragment of the CBFB Break Apart FISH Probe kit labeled with the fluorochrome CytoOrange covers the 5’ (start) portion of the CBFB gene and some adjacent genomic sequences. The 3’ Telomeric fragment of the CBFB Break apart FISH Probe Kit labeled with CytoGreen covers the 3’ (end) part as well as sequences downstream of the gene. The two probes are flanking sequences across the CBFB gene in which variable breakpoints have been observed.
-
پروبهای فیش
پروب فیش CDKN2A/CCP9 FISH Probe
Rated 0 out of 5Read moreINTRODUCTION: CDKN2A, also known as cyclin-dependent kinase Inhibitor 2A, is a gene which in humans is located at chromosome 9, band p21.3. It is ubiquitously expressed in many tissues and cell types. The gene codes for two proteins, including the INK4 family members, p16 (or p16INK4a) and p14arf. Both act as tumor suppressors by regulating the cell cycle. Abnormalities in CDKN2A – also known as ARF, MLM, P14, P16, P19, CMM2, INK4, MTS1, TP16, CDK4I, CDKN2, INK4A, MTS-1, P14ARF, P19ARF, P16INK4, P16INK4A or P16-INK4A – occur in gliomas and meningioma’s as well as numerous other familial and sporadic tumor types.
INTENDED USE: The CDKN2A/CCP9 FISH Probe Kit is designed to detect the human CDKN2A gene located on chromosome band 9p21.3, along with the number of chromosome 9 copies per cell and measure the copy number of the human CDKN2A gene located on chromosome band 9p21.3.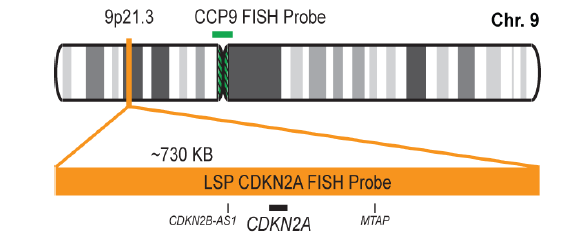
PROBE DESCRIPTION: The CDKN2A Probe of the CDKN2A/CCP9 FISH Probe kit labeled with the fluorochrome CytoOrange covers a chromosomal region which includes the entire CDKN2A gene. The CCP9 Probe of the CDKN2A/CCP9 FISH Probe kit labeled with the flourochrome CytoGreen is derived from chromosome 9-specific alpha satellite DNA is designed to serve as a control to determine the number of chromosome 9 copies per cell.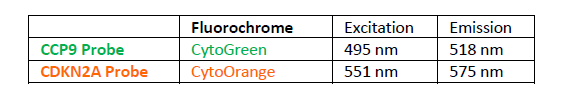
-
پروبهای فیش
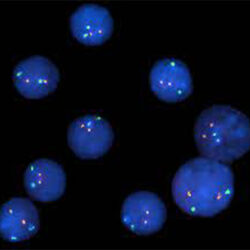
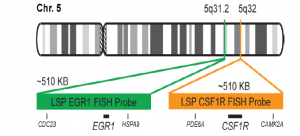
پروب فیش CSF1R/EGR1 FISH Probe
Rated 0 out of 5Read moreINTRODUCTION:The CSF1R gene (known also as c-FMS) is a receptor for a cytokine called colony stimulating factor 1 that in humans encode cell-surface protein called Colony stimulating factor 1 receptor (CSF1R), also known as macrophage colony-stimulating factor receptor (M-CSFR), and CD115 (Cluster of Differentiation 115). he encoded protein is a single pass type I membrane protein and acts as the receptor for colony stimulating factor 1, a cytokine which controls the production, differentiation, and function of macrophages. The EGR1 gene encode in humans a protein called EGR-1 (Early growth response protein 1) also known as Zif268 (zinc finger protein 225) or NGFI-A (nerve growth factor-induced protein A). EGR-1 is a transcription factor. Abnormalities in CSF1R – also known as FMS, CSFR, FIM2, HDLS, CFMS, CD115, CSF1R or M-CSF-R – and abnormalities in EGR1 – also Known as ERBB, HER1, mENA, ERBB1, PIG61 or NISBD2 – have been observed in myeloid malignancies, lung cancer and several other cancer types.
INTENDED USE: The CSF1R/EGR1 FISH Probe Kit is designed to detect the human CSF1R gene, located on chromosome band 5q32, and the EGR1 gene on chromosome band 5q31.2.PROBE DESCRIPTION:The CSF1R Probe of the CSF1R/EGR1 FISH Probe kit labeled with the fluorochrome CytoOrange covers a chromosomal region which includes the entire CSF1R gene. The EGR1 Probe of the CSF1R/EGR1 FISH Probe Kit labeled with CytoGreen covers a chromosomal region which includes the entire EGR1 gene. Both probes are designed to measure the copy number of the human CSF1R and EGR1 gene located on chromosome band 5q32 and 5q31.2, respectively.

-
پروبهای فیش
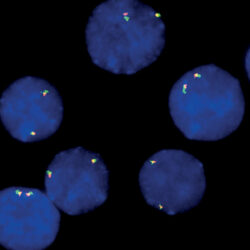
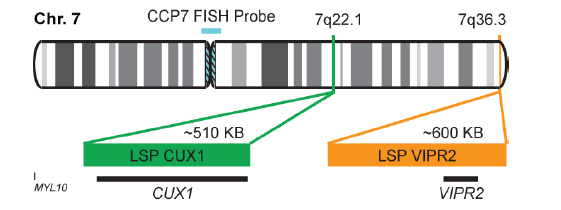
پروب فیش CUX1/VIPR2/CCP7 FISH Probe
Rated 0 out of 5Read moreINTRODUCTION: Expression of the CUX1 gene – also known as CDP, CUX, p75, CASP, CDP1, COY1, Clox, p100, p110, p200, CUTL1, GOLIM6, CDP/Cut, Cux/CDP or Nbla10317 – has been observed elevated in pancreatic, breast and other cancers. Duplications and other anomalies in the region of the VIPR2 gene – also called VPAC2, VPAC2R, VIP-R-2, VPCAP2R, PACAP R3, DUP7q36.3, PACAP-R-3 or C16DUPq36.3 – are associated with schizophrenia, prenatal malformations and some intestinal malignancies.
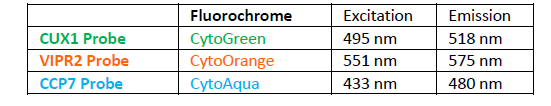
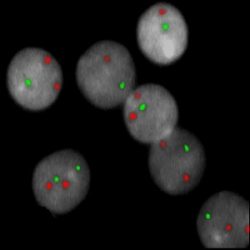
INTENDED USE: The CUX1/VIPR2/CCP7 FISH Probe Kit is designed to detect the human CUX1 gene located on chromosome band 7q22.1, and the VIPR2 gene on chromosome band 7q36.3, along with the number of chromosome 7 copies per cell.PROBE DESCRIPTION: The VIPR2 Probe of the CUX1/VIPR2/CCP7 FISH Probe Kit labeled with the fluorochrome CytoOrange covers a chromosomal region which includes the entire CUX1 gene. The CUX1 Probe of the CUX1/VIPR2/CCP7 FISH Probe Kit labeled with CytoGreen covers a chromosomal region which includes the entire VIPR2 gene. The CCP7 Probe of the CUX1/VIPR2/CCP7 FISH Probe Kit labeled with the fluorochrome CytoAqua is derived from chromosome 7-specific alpha satellite DNA, is designed to serve as a control to determine the number of chromosome 7 copies per cell.
The probes are designed measure the copy number of the human CUX1 and VIPR2 genes located on chromosome bands 7q22.1 and 7q36.3, respectively. -
پروبهای فیش
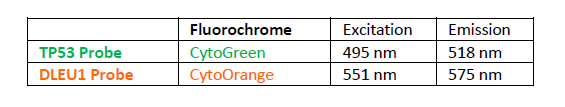
پروب فیش DLEU1/TP53 FISH Probe
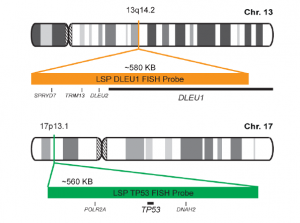
Rated 0 out of 5Read moreINTRODUCTION: The DLEU1 gene was originally identified as a potential tumor suppressor gene. The TP53 gene encodes a tumor suppressor protein containing transcriptional activation, DNA binding, and oligomerization domains. The encoded protein responds to diverse cellular stresses to regulate expression of target genes, thereby inducing cell cycle arrest, apoptosis, senescence, DNA repair, or changes in metabolism. Rearrangements and abnormal expression of the DLEU1 gene region – also known as BCMS, DLB1, LEU1, LEU2, XTP6, BCMS1, DLEU2, LINC00021 or NCRNA00021 – and the TP53 gene – also known as P53, BCC7, LFS1 or TRP53– have been observed in B-cell chronic lymphocytic leukemia (CLL) and other malignancies.
INTENDED USE: The DLEU1/TP53 FISH Probe Kit is designed to detect the human DLEU1 and TP53 genes located on chromosome bands 13q14.2 and 17p13.1, respectively. Both probes are designed to measure the copy number of the human DLEU1 and TP53 genes located on chromosome bands 13q14.2 and 17p13.1, respectively.PROBE DESCRIPTION: The DLEU1 Probe of the DLEU1/TP53 FISH Probe Kit labeled with the fluorochrome CytoOrange covers the center sequences and the 5′ (start) portion of the DLEU1 gene and some adjacent genomic sequences. The TP53 Probe of the DLEU1/TP53 FISH Probe Kit labeled with CytoGreen covers a chromosomal region which includes the entire TP53 gene.
-
پروبهای فیش
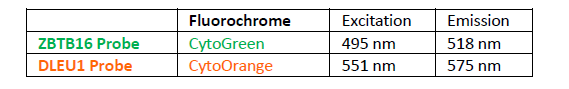
پروب فیش DLEU1/ZBTB16 FISH Probe
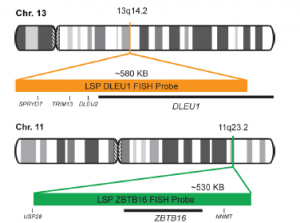
Rated 0 out of 5Read moreINTRODUCTION:The DLEU1 gene was originally identified as a potential tumor suppressor gene.The ZBTB16 gene is a member of the Kruppel C2H2 type zinc finger protein family and encodes a transcription factor containing nine Kruppel-type zinc finger domains at the C-terminus. The ZBTB16 protein is located in the cell nucleus and is involved in the progression of the cell cycle, interacting with histone deacetylases. Rearrangements and abnormal expression of the DLEU1 gene region – also known as BCMS, DLB1, LEU1, LEU2, XTP6, BCMS1, DLEU2, LINC00021 or NCRNA00021 – have been observed in B-cell chronic lymphocytic leukemia (CLL), multiple myeloma (MM) and other malignancies. Fusions and aberrant expression of the ZBTB16 gene – also known as PLZF or ZNF145 – have been reported in acute promyelocytic leukemia and other neoplasias.
INTENDED USE: The DLEU1/ZBTB16 FISH Probe Kit is designed to detect the human DLEU1 gene, located on chromosome band 13q14.2, and the ZBTB16 gene on chromosome band 11q23.2.PROBE DESCRIPTION:The DLEU1 Probe of the DLEU1/ZBTB16 FISH Probe kit labeled with the fluorochrome CytoOrange covers a chromosomal region which includes the majority of the DLEU1 gene and genomic sequences adjacent to the 5′ end of the gene. The ZBTB16 Probe of the DLEU1/ZBTB16 FISH Probe Kit labeled with CytoGreen e covers a chromosomal region which includes the entire ZBTB16 gene. Both probes are designed to measure the copy number of the human DLEU1 and ZBTB16 genes located on chromosome bands 13q14.2 and 11q23.2, respectively.
-
پروبهای فیش
پروب فیش IGH-BCL2 Fusion/Translocation FISH Probe
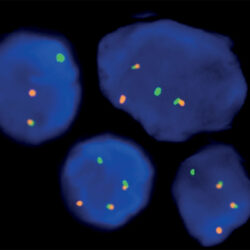
Rated 0 out of 5Read moreINTRODUCTION: Members of the BCL2 family of proteins play important roles in the control of programmed cell death (apoptosis). They comprise proteins that facilitate apoptosis, as well as apoptosis inhibitors. Lymphomas are malignant conditions of the lymphatic part of the circulatory system. One of the most frequent subtypes of lymphoma in adults is follicular lymphoma, a cancer originating in certain B cells that is often symptomless in its early stages. Nearly half of lymphomas diagnosed in adults are of the follicular subtype. One of the widely recognized molecular characteristics of this subtype of the condition is that BCL2 is regularly dysregulated in follicular lymphomas, most often by gene translocation. In fact, BCL2 protein has been detected in virtually all transformed cells in samples from follicular lymphoma patients, whereas it was not seen in non-neoplastic cells or in normal lymph nodes. One particularly common rearrangement is a reciprocal translocation, in which the BCL2 gene, normally on chromosome 18, is relocated to the immunoglobulin heavy chain (IGH) gene region on chromosome 14 and is overexpressed under the influence of an enhancer element on that locus. However, fusion of BCL2 with other genes, including AFF3 and the IGHL@ and IGK@ gene loci, has also been observed.
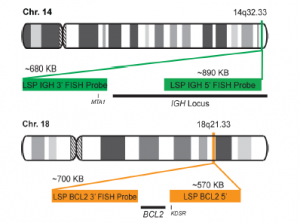
INTENDED USE: The IGH-BCL2 Fusion/Translocation FISH probe kit is designed to detect rearrangements involving regions of the human IGH locus, located on chromosome band 14q32.33, and of the human BCL2 gene on 18q21.33. The kit contains two differentially labeled Locus Specific Probe (LSP) pairs. One set of probes covers the 5’ (start) portion of the BCL2 gene and some upstream untranslated genomic sequence as well as sequences downstream of the 3’ (end) part of the gene. The two probes are flanking sequences on both sides and inside the BCL2 gene where breakpoint clusters and individual breakpoints have been found. The second probe pair spans the immunoglobulin heavy chain gene locus (IGH@) and consists of a portion matching the 5’ (start) region encoding variable gene segments (IGHV), and a part that covers the 3’ (end) and downstream sequences. This probe pair is flanking an area of the IGH gene comprising constant gene segments (IGHC) as well as the delta segment (IGHD), joining region (IGHJ) and epsilon domain (IGHE). Almost all known breakpoints in IGH@have been mapped to this region.
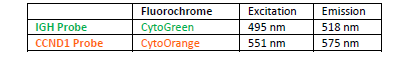
PROBE DESCRIPTION: The BCL2 Probe of the IGH-BCL2 Fusion/Translocation FISH Probe Kit labeled with the fluorochrome CytoOrange covers some genomic sequences adjacent to the 5’ (start) portion of the BCL2 gene; it also covers sequences downstream of the 3’ end of the gene. The IGH Probe of the IGH-BCL2 Fusion/Translocation FISH Probe Kit labeled with the fluorochrome CytoGreen covers the 5’ and the center sequences of the IGH locus, and it also covers the 3’ (end) part and the neighboring downstream region.
-
پروبهای فیش
پروب فیش IGH-CCND1 Fusion-Translocation FISH Probe
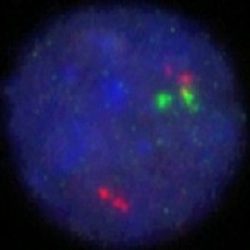
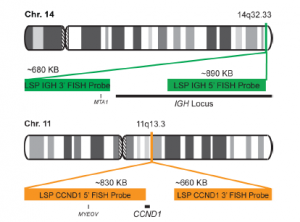
Rated 0 out of 5Read moreINTRODUCTION: Cyclin D1, also called B-cell leukemia/lymphoma 1 (BCL1) or parathyroid adenomatosis 1 (PRAD1), belongs to a family of highly conserved proteins that serve as key regulators of the cell cycle. They form complexes with, and thereby regulate the activity of cyclin-dependent kinases (CDKs), a family of protein kinase enzymes required for progression through the cell cycle. Cyclin D1 is overexpressed in many tumor types. For example, it has been found amplified, and proven to be a marker of poor prognosis, in a percentage of breast cancer cases. CCND1 is also aberrantly activated in a number of hematological malignancies. It is mainly found in mantle cell lymphoma (MCL), a relatively rare non-Hodgkin’s lymphoma type. The most common translocation in MCL is one in which CCND1 is moved to the IGH locus on chromosome 14; juxtaposition of the two genes leads to activation of the CCND1 gene via enhancer elements in the IGH locus. However, fusion of CCND1 with other genes, including FSTL3 and the IGHL@ and IGK@ gene loci, has also been observed. CCND1 activation by translocation also occurs in B-prolymphocytic leukemia, plasma cell leukemia, splenic lymphoma with villous lymphocytes and sometimes in chronic lymphocytic leukemia and multiple myeloma. Detection of CCND1 gene translocation is now widely recommended for the discernment of specific lymphoma subtypes. Furthermore, it is a predictor of improved survival in multiple myeloma patients.
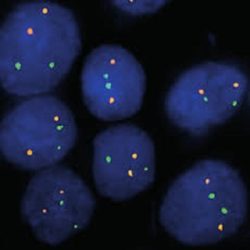
INTENDED USE: The IGH-CCND1 Fusion/Translocation FISH probe kit is designed to detect rearrangements involving regions of the human IGH locus, located on chromosome band 14q32.33, and of the human CCND1 gene on 11q13.3. The kit contains two differentially labeled Locus Specific Probe (LSP) pairs. One set of probes covers the 5’ (start) portion of the CCND1 gene and some upstream untranslated genomic sequence as well as sequences downstream of the 3’ (end) part of the gene. The two probes are flanking sequences in the CCND1gene in which both a 3’ breakpoint cluster as well as several individual breakpoints have been observed.PROBE DESCRIPTION: The IGH Probe of the IGH-CCND1 Fusion/Translocation FISH Probe Kit labeled with the fluorochrome CytoGreen covers the 5’ and the center sequences of the IGH locus, and it also covers the 3’ (end) part and the neighboring downstream region. The CCND1 Probe of the IGH-CCND1 Fusion/Translocation FISH Probe Kit labeled with the fluorochrome CytoOrange covers some genomic sequences adjacent to the 5’ (start) portion of the CCND1 gene; it also covers sequences downstream of the 3’ end of the gene. The probe set is optimized to reveal translocations between the two regions.
-
پروبهای فیش
پروب فیش PML-RARA Fusion/Translocation FISH Probe
Rated 0 out of 5Read moreINTRODUCTION:PML gene encodes in humans a protein calledPromyelocytic leukemia protein (PML) (also known as MYL, RNF71, PP8675 or TRIM19). PML protein is a tumor suppressor protein required for the assembly of a number of nuclear structures, called PML-nuclear bodies, which form amongst the chromatinof the cell nucleus. The RARA gene encodes in humans a nuclear receptor protein called Retinoic acid receptor alpha (RAR-α), also known as NR1B1 (nuclear receptor subfamily 1, group B, member 1). Rearrangements between the two genes have been observed in acute promyelocytic leukemia and other malignancies.
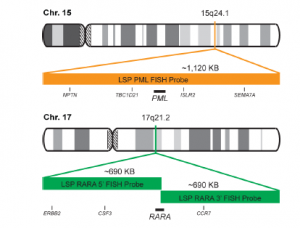
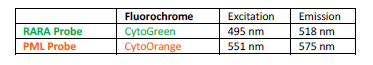
INTENDED USE: The PML-RARA Fusion/Translocation FISH Probe Kit is designed to detect rearrangements involving the human PML and RARA genes, located on chromosome bands 15q24.1 and 17q21.2, respectively.PROBE DESCRIPTION: The RARA Probe of the PML-RARA Fusion/Translocation FISH Probe labeled with the fluorochrome CytoGreen overs a chromosomal region including the entire RARA gene and some sequences upstream (5′ start) and downstream (3′ end) of the gene. The PML Probe of the PML-RARA
Fusion/Translocation FISH Probe labeled with the fluorochrome CytoOrange covers a chromosomal region which includes the entire PML gene and some
sequences around the gene. -
پروبهای فیش
پروب فیش IGH-MAF Fusion/Translocation FISH Probe
Rated 0 out of 5Read moreINTRODUCTION: MAF gene encode in humanas the Transcription factor Maf also known as proto-oncogene c-Maf or V-maf musculoaponeurotic fibrosarcoma oncogene homolog. t(14;16)(q32;q23) represents 14q32/IGH rearrangement and belongs to the group of IGH/MAF translocations (rearrangements of the genes from the MAF oncogene family MAF, MAFA and MAFB with the IGH locus). As the other two IGH/MAF translocations t(8;14)q24;q32) andt(14;20)(q13;32) is described only in plasma cell neoplasms (PCN). t(14;16)(q32;q23) resulted in the juxtaposition of the oncogene MAF ( located at 16q23) to the strong enhancer of the IGH gene (located at 14q32) causing its up regulation in the plasma cells. This anomaly is found in both multiple myeloma (MM) and its precursor monoclonal gammapathy of undetermined significance (MGUS) respectively in 5% and 1-5% of the cases with 14q32 rearrangements.
INTENDED USE: The IGH-MAF Fusion/Translocation FISH probe kit is designed to detect rearrangements involving regions of the human IGH locus, located on chromosome band 14q32.33, and of the human MAF gene on 16q23.2.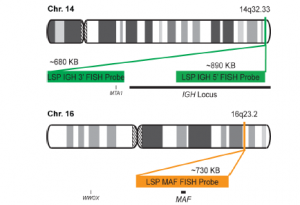
PROBE DESCRIPTION: The IGH Probe of the IGH-MAF Fusion/Translocation FISH Probe Kit labeled with the fluorochrome CytoGreen covers the 5’ and the center sequences of the IGH locus, and it also covers the 3’ (end) part and the neighboring downstream region. The MAF Probe of the IGH-MAF Fusion/Translocation FISH Probe Kit labeled with the fluorochrome CytoOrange covers the chromosomal region that includes the entire MAF gene. -
پروبهای فیش
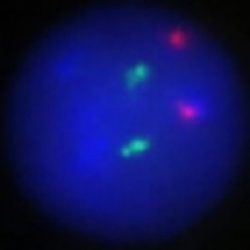
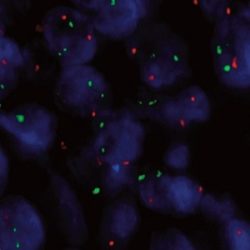
پروب فیش IGH-MYC Fusion/Translocation FISH Probe
Rated 0 out of 5Read moreINTRODUCTION: Rearrangements and abnormal expression of the MYC gene (also known as MRTL, MYCC, c-Myc or bHLHe39) have been described in Burkitt’s lymphoma, where its positivity has diagnostic utility within a clinical, morphological and immunohistochemical context. On the other hand, in large or non-classifiable diffuse B-cell lymphoma, the MYC translocation has a prognostic value and dictates a more aggressive treatment of the neoplasm. On the other hand, together with the determination of the translocation of the BCL2 and BCL6 genes, it aims to establish the double or triple-hit character of the neoplasm. Other hematological neoplasms, myelomas, as well as breast, cervical, colon, ovarian and other tumors may present MYC translocation.
Most Burkitt lymphoma cases show the t(8;14)(q24;q32) [MYC-IGH] and less commonly the t(8;22)(q24;q11) or t(2;8)(p12;q24). The translocation is present in both the endemic African Burkitt lymphoma and in the non-endemic tumor type (Europe, America, and Japan). In case the Burkitt lymphoma infiltrated the bone marrow (leukemic phase) the MYC-translocation can be demonstrated in the bone marrow or blood as well.
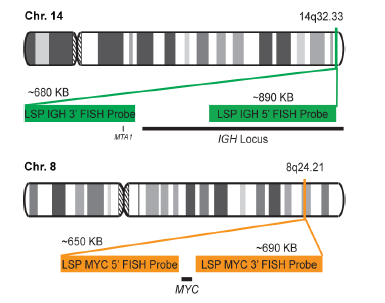
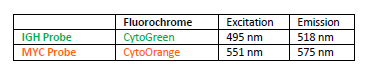
INTENDED USE: The IGH-MYC Fusion/Translocation FISH probe kit t is designed to detect rearrangements involving the human IGH locus and MYC gene, located on chromosome bands 14q32.33 and 8q24.21, respectively.PROBE DESCRIPTION: The IGH Probe of the IGH-MYC Fusion/Translocation FISH Probe Kit labeled with the fluorochrome CytoGreen covers the 5’ and the center sequences of the IGH locus, and it also covers the 3’ (end) part and the neighboring downstream region. The MYC Probe of the IGH-MYC Fusion/Translocation FISH Probe Kit labeled with the fluorochrome CytoOrange covers some genomic sequences downstream of the 5’ (start) portion of the MYC gene, and it also covers the sequences adjacent to the 3’ end of the gene. The probe set is optimized to reveal translocations between the two regions.
-
پروبهای فیش
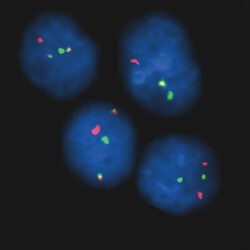
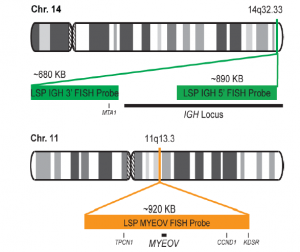
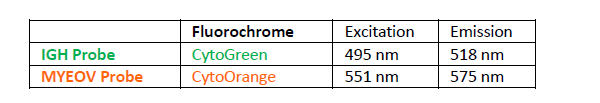
پروب فیش IGH-MYEOV Fusion/Translocation FISH Probe
Rated 0 out of 5Read moreINTRODUCTION: The MYEOV gene was originally isolated by the application of the NIH/3T3 tumorigenicity assay with DNA from a gastric carcinoma. The chromosomal region 11q13 is frequently associated with genetic rearrangements in a large number of human malignancies, including B-cell malignancies and overexpression of MYEOV is frequently observed in breast tumors and oral, esophageal squamous cell carcinomas and multiple myeloma. MYEOV is a putative oncogene, located 360kb away from CCND1, which is thought to be activated in the translocation by becoming closely associated with IGH enhancers. The mantle cell lymphoma (MCL) is a B-cell non-Hodgkin lymphoma with an aggressive clinical course. It is genetically characterized by t(11;14)(q13;q32) and is present in about 95% of MCL patients. By lower frequency, t(11;14) is also detectable in B-cell prolymphocytic leukemia, myelomas and chronic lymphocytic leukemia. In multiple myeloma (MM), t(11;14) is the most common translocation, detectable in about 15-20% of all MM patients by FISH.
INTENDED USE: The IGH-MYEOV Fusion/Translocation FISH probe kit is designed to detect rearrangements involving the human IGH locus and MYEOV gene located on chromosome bands 14q32.33 and 11q13.3, respectively.PROBE DESCRIPTION:The IGH Probe of the IGH-MYEOV Fusion/Translocation FISH Probe Kit labeled with the fluorochrome CytoGreen covers the 5’ and the center sequences of the IGH locus; it also covers the 3’ part and the neighboring downstream region. The MYEOV Probe of the IGH-MYEOV Fusion/Translocation FISH Probe Kit labeled with the fluorochrome CytoOrange covers a chromosomal region which includes the entire MYEOV gene. The probe set is optimized to reveal translocations between the two regions.